|
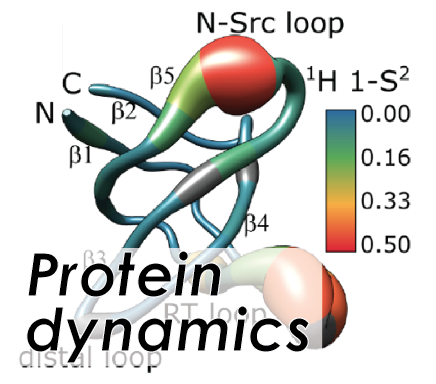
Structural biology is concerned with elucidation of biomolecules, in particular proteins, with respect to their atomic-resolution 3D structures, dynamics, and interactions with other molecules in order to shine light on the principles behind their physiological functionality. As such, the related research yields a detailed understanding of the mechanisms constituting cellular life and the possibilities of medical treatment to interfere with any aberrant or mis-regulated processes. In particular, the dynamics of proteins, happening on manifold timescales from ps to ms, represent a feature that has been difficult to grasp but constitutively important for the individual proteins' functionality.
Nuclear Magnetic Resonance (NMR) spectroscopy is based on the interaction of nuclear magnetic momenta with an external magnetic field and the readout of resonance frequencies, lifetimes of non-equilibrium states, and spatial spin proximities. Employing such principles, the technique is capable of assessing molecular structure and dynamics with of atomic resolution. This can be applied to proteins: Soluble proteins can be characterized using a huge set of established solution NMR methodology. On the other hand, solid-state NMR is applied for example to study membrane proteins in a lipid-bilayer setting or amyloidogenic proteins. This approach has witnessed significant developments in the strategies employed in the last years. One such development is the availability of proton detection rather than detection of heteronuclear signals, which has been driven by the methods development for fast Magic-Angle Spinning (MAS) of the sample (on the order of 100 000 rotations per second) in the magnetic field.
Our group focuses on various aspects of NMR spectroscopy, both in terms of methods development as well as application to gain understanding of biology-related questions, and has been able to significantly drive the available methodology and as such the power of NMR in general forward. As such, we strive for methods enabling structure elucidation of proteins with increasing molecular size or which are characterized by significant sample heterogeneity, complex intermolecular interactions, or limitations in isotope-labeled protein expression. This concerns the structure of such proteins as well as the dynamics relevant for protein functionality. We develop new spectroscopic approaches as building blocks of our spectroscopic pulse schemes in conjunction with dedicated data processing schemes. Based on the developed methodology, we are in the position to give answers to questions about the details of specific proteins’ structure and dynamics, protein-water interactions, protein-ligand interactions, ligand mobility, and chemical mechanisms comprised in the active site.
See our (overview) Accounts paper on "Protons in Solid-State NMR"!
|
|