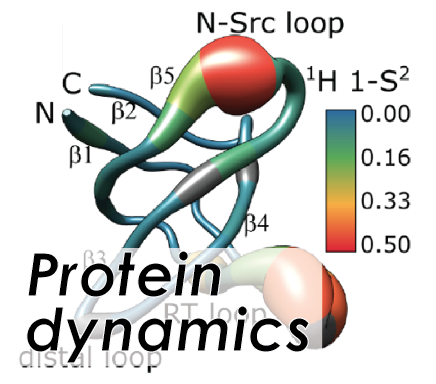
Protein dynamics
Protein dynamics play a huge role for the physiological functionality of proteins, for example for interactions with other proteins or for processing of substrates upon catalysis. NMR is uniquely capable of determining protein dynamics, but new perspectives for improving existing methodology have continuously been arising. Solid-state NMR can determine local motion with high sensitivity over many orders of timescales. Our interest is to expand the set of methods, particularly with regard to the regime of conformational exchange, by taking into account new strategies, as well as to understand the functional dynamics of proteins in the context of protein-protein or protein-small-molecule interactions.
Past and recent research topics in this direction are, for example:
|
Higher-dimensionality dynamics approaches for bigger molecules |
|
|
|
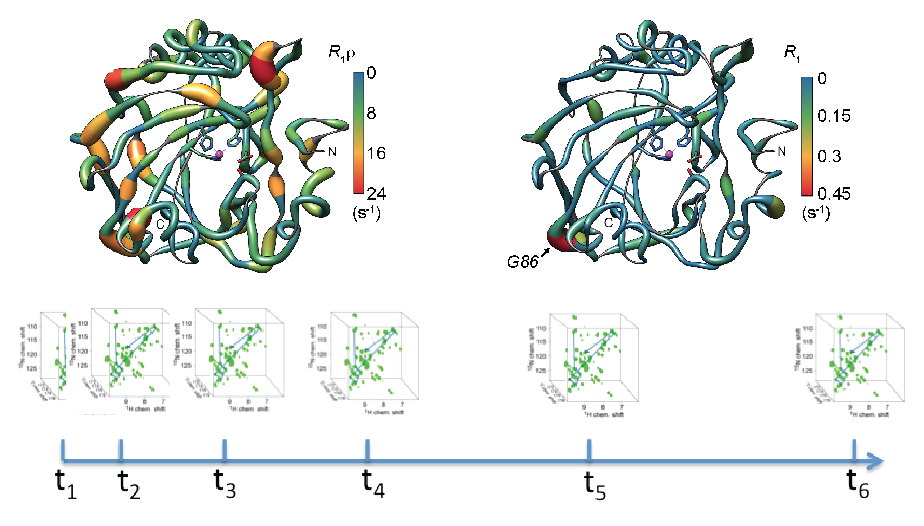
S. K. Vasa, H. Singh, P. Rovó, R. Linser, “Dynamics and interactions of a 29-kDa human enzyme studied by solid-state NMR”, J. Phys. Chem. Lett., 9, 1307−1311 (2018). (Link) |
|
Protons as reporters on dynamics |
|
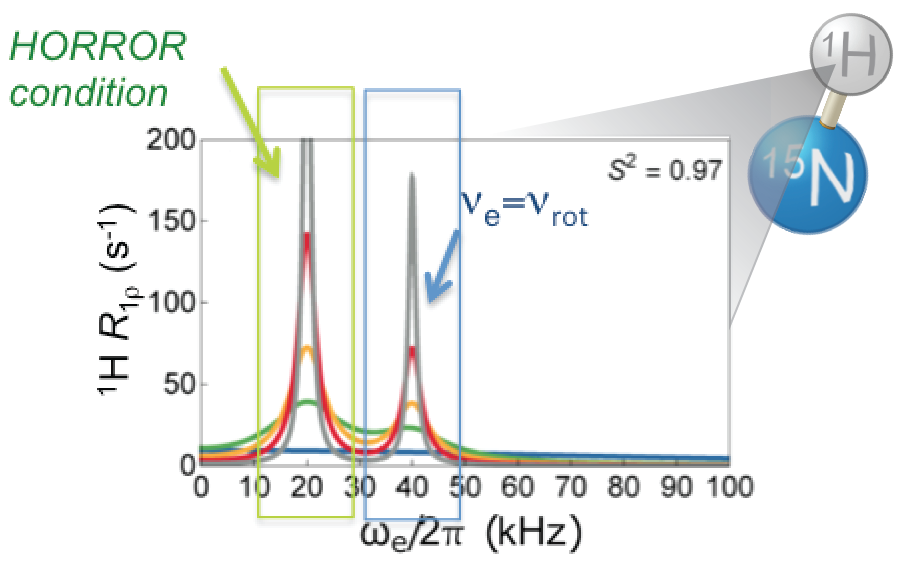 |
P. Rovó, R. Linser, “Microsecond timescale proton rotating-frame relaxation under magic angle spinning”, J. Phys Chem. 121 (25), 6117–6130 (2017). P. Rovó, C. A. Smith, D. Gauto, B. L. de Groot, P. Schanda, R. Linser, “Mechanistic insights into microsecond timescale motion of solid proteins using complementary 15N and 1H relaxation dispersion techniques”, J. Am. Chem. Soc., DOI: 10.1021/jacs.8b09258 (2019). (Link) |
| New approaches for gathering µs dynamics data | |
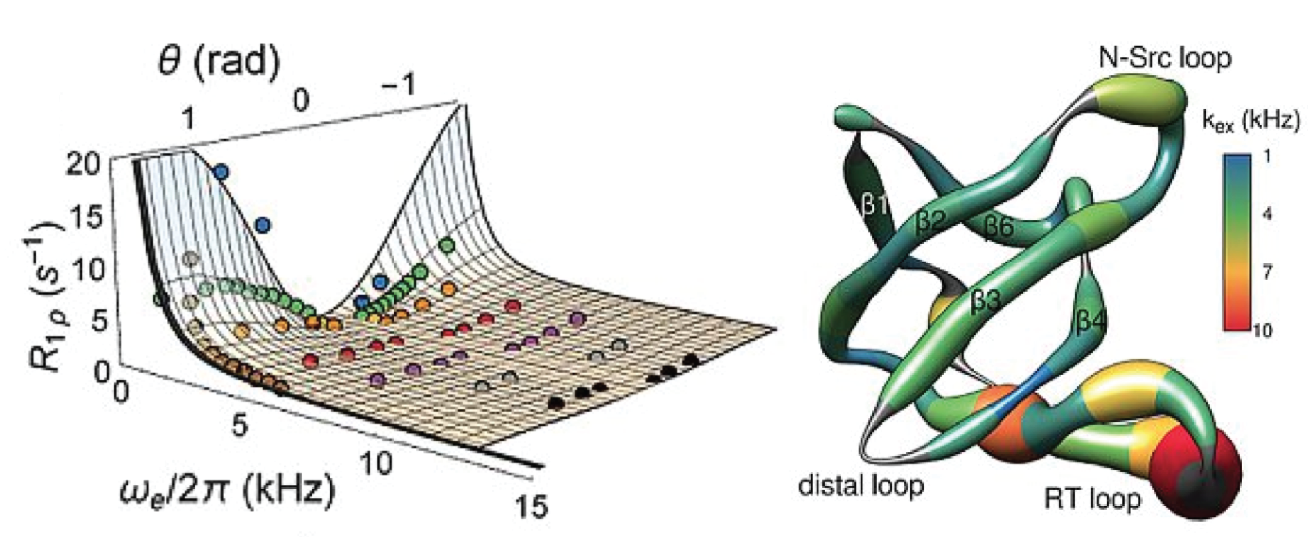 |
R. Linser, U. Fink, B. Reif, “Detection of dynamic regions in biological solids enabled by spin-state selective NMR experiments”, J. Am. Chem. Soc. 132 (26), 8891–8893 (2010). R. Linser, R. Sarkar, A. Krushelnitzky, A. Mainz, B. Reif, “Dynamics in the Solid State: Perspectives for the Investigation of Amyloid Aggregates, Membrane Proteins, and Soluble Protein Complexes”, J. Biomol. NMR 59 (1), 1–14 (2014). P. Rovó, R. Linser, “Microsecond timescale solid-state backbone dynamics: a combined NMR approach”, ChemPhysChem, 19, 34–39 (2018). K. Grohe, S. Patel, C. Hebrank, S. Medina, A. Klein, P. Rovó, S. K. Vasa, H. Singh, B. Vögeli, L. V. Schäfer, R. Linser, “Protein motional details revealed by complementary structural-biology techniques”, Structure (Cell Press), 28, 1-11, DOI: 10.1016/j.str.2020.06.001 (2020). (Link) |
|
Dynamics applications |
|
 |
R. Linser, N. Salvi, R. Briones, P. Rovó, B. de Groot, G. Wagner, “The Membrane Anchor of the Transcriptional Activator SREBP is Characterized by Intrinsic Conformational Flexibility”, Proc. Natl. Acad. Sci. USA, 112 (40) 12390-12395 (2015). |
 |
H. Singh, S. K. Vasa, H. Jangra, P. Rovó, C. Päslack, C. K. Das, H. Zipse, L. Schäfer, R. Linser, “Fast-microsecond dynamics of the protein-water network in the active site of human carbonic anhydrase II by solid-state NMR spectroscopy.” J. Am. Chem. Soc., 141 (49), 19276-19288, DOI: 10.1021/jacs.9b05311 (2019). (Link) (Cover article: See the picture here!)
|
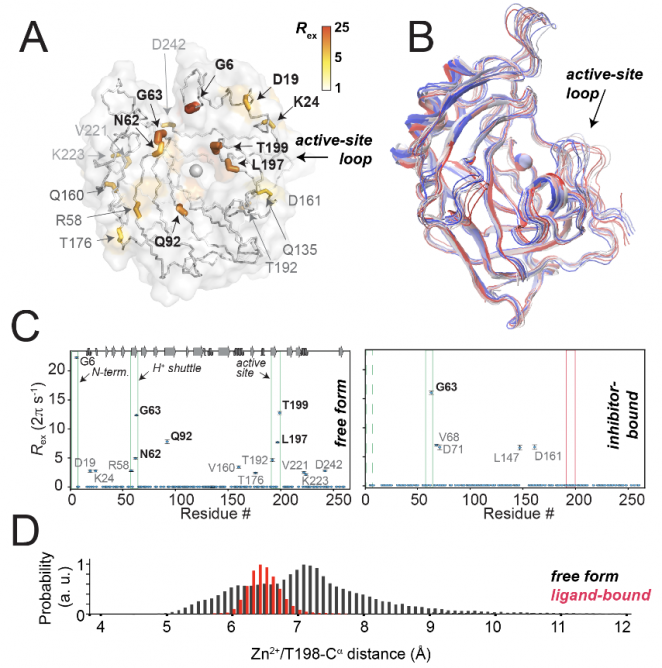 |
H. Singh, C. K. Das S. K. Vasa, K. Grohe, L. V. Schäfer, R. Linser, “The active site of a prototypical “rigid” drug target is marked by extensive conformational dynamics”, Angew. Chem., Int. Ed., 59 (51), 22916-22921(2020), DOI: 10.1002/anie.202009348 (Link).
|